| 出品公司: | ZYbscience |
|---|---|
| 载体名称: | pRetroQ-AcGFP1-N1 |
| 质粒类型: | 逆病毒载体;荧光报告载体 |
| 高拷贝/低拷贝: | 低拷贝 |
| 克隆方法: | 限制性内切酶,多克隆位点 |
| 启动子: | CMV IE |
| 载体大小: | 7606 bp |
| 5' 测序引物及序列: | -- |
| 3' 测序引物及序列: | -- |
| 载体标签: | AcGFP1(C-端) |
| 载体抗性: | 氨苄青霉素 |
| 筛选标记: | 嘌呤霉素(Puromycin) |
| 克隆菌株: | DH5α |
| 宿主细胞(系): | 常规细胞系(293、CV-1、CHO等) |
| 备注: | pRetroQ-AcGFP1-N1载体是自我失活型逆病毒载体,表达C端AcGFP1融合蛋白; pRetroQ-AcGFP1-N1载体能够高效递送目的基因到靶细胞中,尤其适用于原代细胞和其它难转染的细胞系。 |
| 产品目录号: | ZY632505 |
| 稳定性: | 稳表达 |
| 组成型/诱导型: | 组成型 |
| 病毒/非病毒: | 逆转录病毒 |

产品详情
文献和实验
相关推荐
保存条件 :-20℃低温保存
保质期 :三年
英文名 :pRetroQ-AcGFP1-N1
库存 :20
供应商 :泽叶生物
载体基本信息
载体质粒图谱和多克隆位点信息
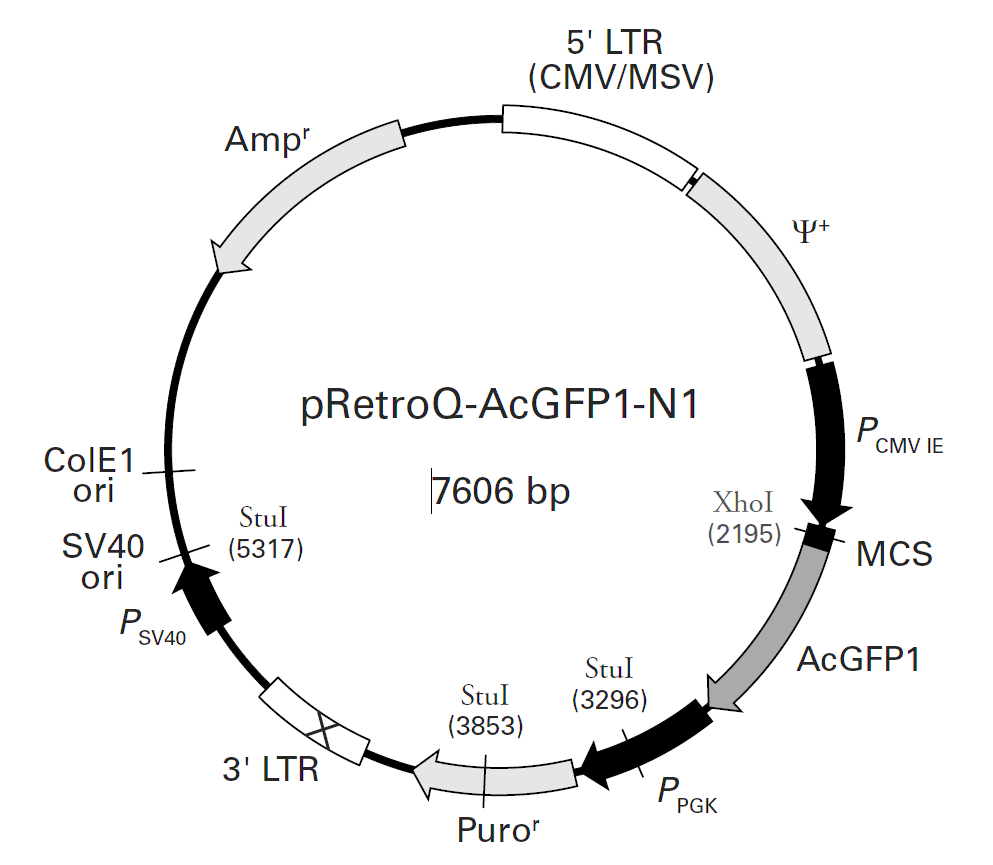
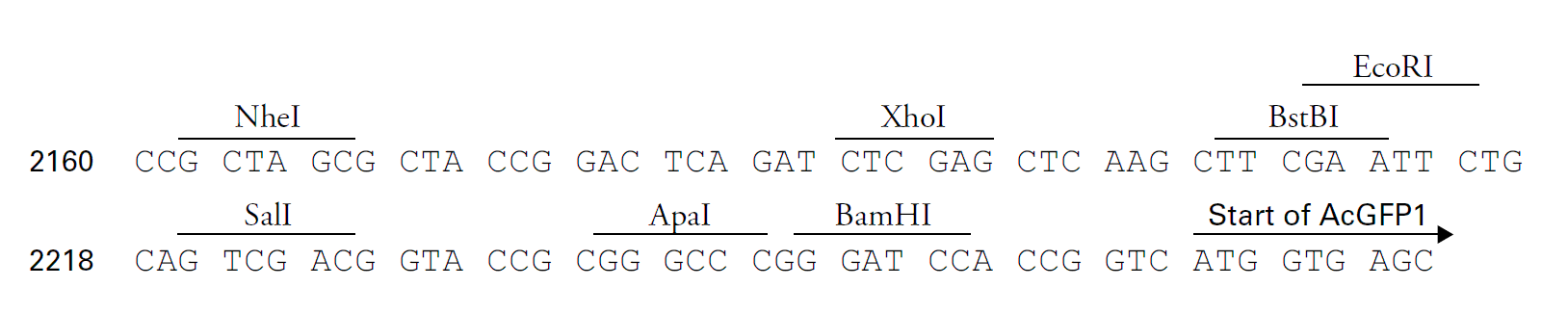
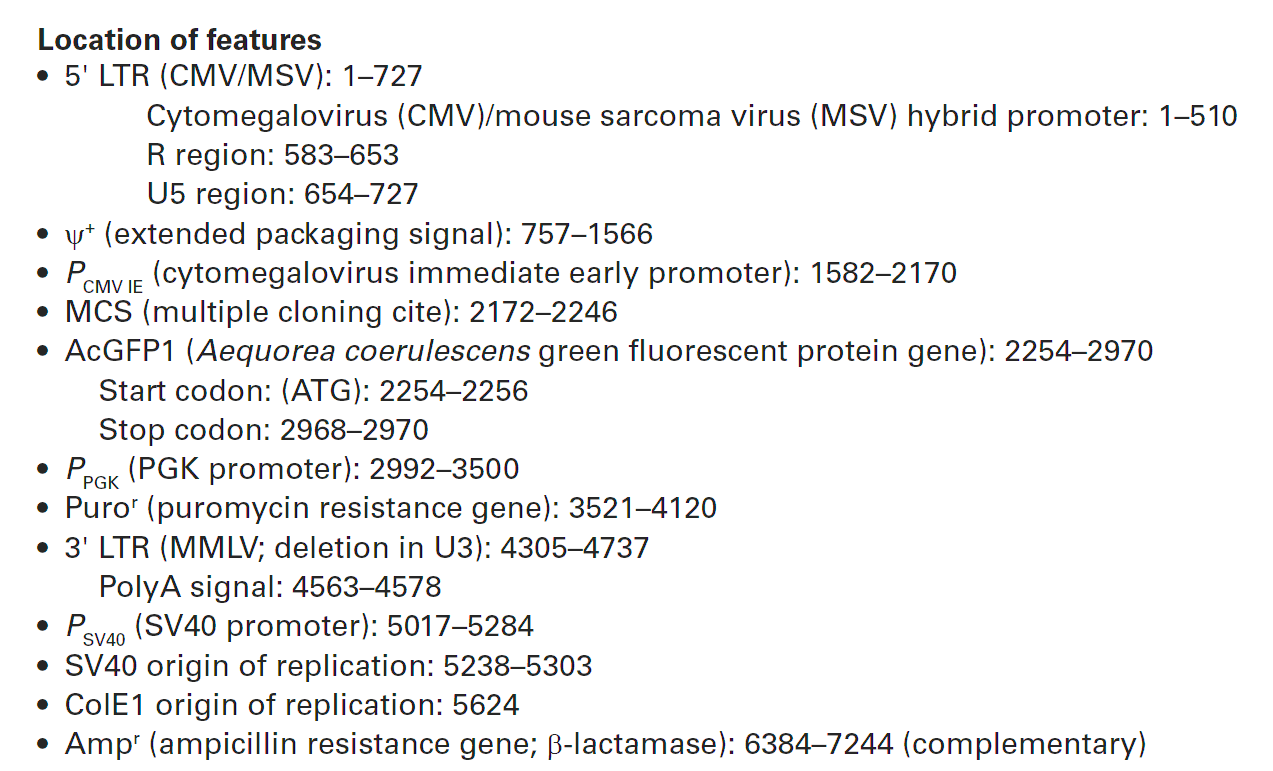
载体简介
pRetroQ-AcGFP1-N1载体描述pRetroQ-AcGFP1-N1 is a high-titer, self-inactivating retroviral vector that facilitates efficient delivery and expression of AcGFP1, as well as N terminal fusions of AcGFP1, to target cells. AcGFP1 is the green fluorescent protein from Aequorea coerulescens (AcGFP1; Excitation maximum = 475 nm; emission maximum = 505 nm).The coding sequence of the AcGFP1 gene contains silent base changes, which correspond to human codon-usage preferences (1). The MCS in pRetroQAcGFP1-N1 lies between the immediate early promoter of CMV (PCMV IE) and the AcGFP1 coding sequences. Genes cloned into the MCS are expressed as fusions to the N-terminus of AcGFP1 when they are in the same reading frame as AcGFP1 and there are no intervening stop codons.The RetroQ retroviral vector backbone incorporates several unique features. This vector contains a puromycin resistance cassette (Puror) driven by the PGK promoter (PPGK) for selection of positively-infected cells (2).The hybrid 5’ long terminal repeat (LTR) consists of the CMV type I enhancer and the murine sarcoma virus (MSV) promoter. This vector demonstrates high levels of transcription in HEK 293-based packaging cell lines due, in part, to the presence of adenoviral E1A (3–6) in these cells. The self-inactivating feature of the vector is provided by a deletion in the 3’ LTR enhancer region (U3). During reverse transcription of the retroviral RNA, the inactivated 3’ LTR is copied and replaces the 5’ LTR CMV enhancer sequences. This can reduce the phenomenon known as promoter interference (7) and allow more efficient expression.Additionally, the viral genomic transcript contains the necessary viral RNA processing elements, including the LTRs, packaging signal (ψ+), and tRNA primer binding site. pRetroQ-AcGFP1-N1 contains a bacterial origin of replication, an E. coli Ampr gene for propagation and selection in bacteria, and an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen.pRetroQ-AcGFP1-N1 is designed to efficiently deliver and express fusions to the N terminus of AcGFP1 into primary cells or cells that are difficult to transfect. Fusions to the N terminus of AcGFP1 retain the fluorescent properties of the native protein, allowing the in vivo localization of the fusion protein. The target gene should be cloned into pRetroQ AcGFP1-N1 so that it is in frame with the AcGFP1 coding sequences, with no intervening in-frame stop codons. The inserted gene should include the initiating ATG codon. The recombinant AcGFP1 vector can be infected or transfected into mammalian cells. If required, stable transformants can be selected using puromycin. pRetroQ-AcGFP1-N1 can also be used simply to express AcGFP1 in a cell line of interest (e.g., as an infection marker).Once pRetroQ-AcGFP1-N1 is transfected into a packaging cell line (such as the RetroPack PT67 Cell line, AmphoPack-293, EcoPack2-293, Pantropic Expression System, or Retro-X Universal Packaging System , RNA from the vector is packaged into non-infectious, replication-incompetent retroviral particles, since pRetroQ-AcGFP1-N1 lacks the structural genes (gag, pol, and env) necessary for particle formation and replication. These genes, however, are stably integrated as part of the packaging cell genome. Once a high-titer supernatant is produced, these retroviral particles can infect target cells and transmit the gene of interest but cannot replicate within these cells due to the absence of viral structural genes. The separate introduction and integration of the structural genes into the packaging cell line minimizes the chances of producing replication-competent virus due to recombination events during cell proliferation.Propagation in E. coli Suitable host strains: DH5α, Fusion Blue, and other general-purpose strains. Selectable marker: plasmid confers resistance to ampicillin (100 μg/ml) in E. coli hosts. E. coli replication origin: ColE1 Copy number: low
载体序列
LOCUS pRetroQ-AcGFP1-N1 7606 bp DNA SYNDEFINITION pRetroQ-AcGFP1-N1ACCESSION KEYWORDS SOURCE ORGANISM other sequences; artificial sequences; vectors.FEATURES Location/Qualifiers source 1..7606 /organism="pRetroQ-AcGFP1-N1" /mol_type="other DNA" promoter 2..506 /label="CMV_immearly_promoter" misc_feature 6..688 /label="5_LTR" misc_feature 9..296 /label="CAG_enhancer" misc_feature 463..483 /label="CMV_fwd_primer" misc_feature 758..1567 /label="psi_plus_pack" misc_feature 1154..1561 /label="gag" misc_feature 1497..1519 /label="MSCV_primer" misc_feature 1497..1519 /label="pLXSN_5_primer" misc_feature 1535..1551 /label="pBABE_5_primer" promoter 1591..2143 /label="CMV_immearly_promoter" misc_feature 1646..1933 /label="CAG_enhancer" misc_feature 2100..2120 /label="CMV_fwd_primer" promoter 2101..2170 /label="CMV_promoter" CDS 2254..2973 /label="ORF frame 1" /translation="MVSKGAELFTGIVPILIELNGDVNGHKFSVSGEGEGDATYGKLT LKFICTTGKLPVPWPTLVTTLSYGVQCFSRYPDHMKQHDFFKSAMPEGYIQERTIFFE DDGNYKSRAEVKFEGDTLVNRIELTGTDFKEDGNILGNKMEYNYNAHNVYIMTDKAKN GIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDH MIYFGFVTAAAITHGMDELYK*" gene 2257..2970 /label="AcGFP1" /gene="AcGFP1" /translation="MVSKGAELFTGIVPILIELNGDVNGHKFSVSGEGEGDATYGKLT LKFICTTGKLPVPWPTLVTTLSYGVQCFSRYPDHMKQHDFFKSAMPEGYIQERTIFFE DDGNYKSRAEVKFEGDTLVNRIELTGTDFKEDGNILGNKMEYNYNAHNVYIMTDKAKN GIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDH MIYFGFVTAAAITHGMDELYK*" misc_feature complement(3036..3059) /label="MSCV_rev_primer" /translation="MVSKGAELFTGIVPILIELNGDVNGHKFSVSGEGEGDATYGKLT LKFICTTGKLPVPWPTLVTTLSYGVQCFSRYPDHMKQHDFFKSAMPEGYIQERTIFFE DDGNYKSRAEVKFEGDTLVNRIELTGTDFKEDGNILGNKMEYNYNAHNVYIMTDKAKN GIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDH MIYFGFVTAAAITHGMDELYK*" CDS 3281..4120 /label="ORF frame 2" /translation="MEAGRPLGQRPIAALLLRFLGSEAGKGWVRGRAQGRAQGRGGRP KVLRRPGILHASKAHVCRAVLLFLISGPFDLQPKLTMTEYKPTVRLATRDDVPRAVRT LAAAFADYPATRHTVDPDRHIERVTELQELFLTRVGLDIGKVWVADDGAAVAVWTTPE SVEAGAVFAEIGPRMAELSGSRLAAQQQMEGLLAPHRPKEPAWFLATVGVSPDHQGKG LGSAVVLPGVEAAERAGVPAFLETSAPRNLPFYERLGFTVTADVEVPEGPRTWCMTRK PGA*" misc_feature 3433..3452 /label="mPGK_F_primer" /translation="MEAGRPLGQRPIAALLLRFLGSEAGKGWVRGRAQGRAQGRGGRP KVLRRPGILHASKAHVCRAVLLFLISGPFDLQPKLTMTEYKPTVRLATRDDVPRAVRT LAAAFADYPATRHTVDPDRHIERVTELQELFLTRVGLDIGKVWVADDGAAVAVWTTPE SVEAGAVFAEIGPRMAELSGSRLAAQQQMEGLLAPHRPKEPAWFLATVGVSPDHQGKG LGSAVVLPGVEAAERAGVPAFLETSAPRNLPFYERLGFTVTADVEVPEGPRTWCMTRK PGA*" CDS complement(3446..4171) /label="ORF frame 1" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" gene 3521..4120 /label="puro (variant)" /gene="puro (variant)" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" misc_feature complement(4840..4862) /label="pGEX_3_primer" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" misc_feature complement(4998..5018) /label="pBABE_3_primer" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" misc_feature complement(5004..5219) /label="SV40_enhancer" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" promoter 5016..5284 /label="SV40_promoter" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" rep_origin 5183..5260 /label="SV40_origin" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" misc_feature 5245..5264 /label="SV40pro_F_primer" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" gene complement(6384..7244) /label="Ampicillin" /gene="Ampicillin" /translation="MGSCAPFGRALRVVGRASGTGLAGHAPGARSFGHLDVGGDGEAE PLVEGEVAGRGGLQEGGHPGALGRLHSGEHDGAAQTLALVVGRDADGGQEPRGLLGPV RRQEAFHLLLRGQPGTAQLGHARADLGEHRPRFDALRRGPDRHRGAVVRDPHLADVEP DAREEEFLQLGDPLDVAVRIDGVARGGVVGERGGEGAYGPGDVVAGGEAHRGLVLGHG KLGLQVERPGDEEEENSAADVRF*" CDS complement(6384..7244) /label="ORF frame 3" /translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGY IELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRVDAGQEQLGRRIHYSQNDLVE YSPVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRL DRWEPELNEAIPNDERDTTMPAAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPL LRSALPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIA EIGASLIKHW*" promoter complement(7286..7314) /label="AmpR_promoter" /translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGY IELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRVDAGQEQLGRRIHYSQNDLVE YSPVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRL DRWEPELNEAIPNDERDTTMPAAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPL LRSALPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIA EIGASLIKHW*" promoter 7536..506 /label="CMV_immearly_promoter" /translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGY IELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRVDAGQEQLGRRIHYSQNDLVE YSPVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRL DRWEPELNEAIPNDERDTTMPAAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPL LRSALPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIA EIGASLIKHW*" misc_feature 7600..688 /label="5_LTR" /translation="MSIQHFRVALIPFFAAFCLPVFAHPETLVKVKDAEDQLGARVGY IELDLNSGKILESFRPEERFPMMSTFKVLLCGAVLSRVDAGQEQLGRRIHYSQNDLVE YSPVTEKHLTDGMTVRELCSAAITMSDNTAANLLLTTIGGPKELTAFLHNMGDHVTRL DRWEPELNEAIPNDERDTTMPAAMATTLRKLLTGELLTLASRQQLIDWMEADKVAGPL LRSALPAGWFIADKSGAGERGSRGIIAALGPDGKPSRIVVIYTTGSQATMDERNRQIA EIGASLIKHW*"ORIGIN 1 TCCGCGTTAC ATAACTTACG GTAAATGGCC CGCCTGGCTG ACCGCCCAAC GACCCCCGCC 61 CATTGACGTC AATAATGACG TATGTTCCCA TAGTAACGCC AATAGGGACT TTCCATTGAC 121 GTCAATGGGT GGAGTATTTA CGGTAAACTG CCCACTTGGC AGTACATCAA GTGTATCATA 181 TGCCAAGTAC GCCCCCTATT GACGTCAATG ACGGTAAATG GCCCGCCTGG CATTATGCCC 241 AGTACATGAC CTTATGGGAC TTTCCTACTT GGCAGTACAT CTACGTATTA GTCATCGCTA 301 TTACCATGGT GATGCGGTTT TGGCAGTACA TCAATGGGCG TGGATAGCGG TTTGACTCAC 361 GGGGATTTCC AAGTCTCCAC CCCATTGACG TCAATGGGAG TTTGTTTTGG CACCAAAATC 421 AACGGGACTT TCCAAAATGT CGTAACAACT CCGCCCCATT GACGCAAATG GGCGGTAGGC 481 GTGTACGGTG GGAGGTCTAT ATAAGCAGAG CTCAATAAAA GAGCCCACAA CCCCTCACTC 541 GGCGCGCCAG TCTTCCGATA GACTGCGTCG CCCGGGTACC CGTATTCCCA ATAAAGCCTC 601 TTGCTGTTTG CATCCGAATC GTGGTCTCGC TGTTCCTTGG GAGGGTCTCC TCTGAGTGAT 661 TGACTACCCA CGACGGGGGT CTTTCATTTG GGGGCTCGTC CGGGATTTGG AGACCCCTGC 721 CCAGGGACCA CCGACCCACC ACCGGGAGGT AAGCTGGCCA GCAACTTATC TGTGTCTGTC 781 CGATTGTCTA GTGTCTATGT TTGATGTTAT GCGCCTGCGT CTGTACTAGT TAGCTAACTA 841 GCTCTGTATC TGGCGGACCC GTGGTGGAAC TGACGAGTTC TGAACACCCG GCCGCAACCC 901 TGGGAGACGT CCCAGGGACT TTGGGGGCCG TTTTTGTGGC CCGACCTGAG GAAGGGAGTC 961 GATGTGGAAT CCGACCCCGT CAGGATATGT GGTTCTGGTA GGAGACGAGA ACCTAAAACA 1021 GTTCCCGCCT CCGTCTGAAT TTTTGCTTTC GGTTTGGAAC CGAAGCCGCG CGTCTTGTCT 1081 GCTGCAGCGC TGCAGCATCG TTCTGTGTTG TCTCTGTCTG ACTGTGTTTC TGTATTTGTC 1141 TGAAAATTAG GGCCAGACTG TTACCACTCC CTTAAGTTTG ACCTTAGGTC ACTGGAAAGA 1201 TGTCGAGCGG ATCGCTCACA ACCAGTCGGT AGATGTCAAG AAGAGACGTT GGGTTACCTT 1261 CTGCTCTGCA GAATGGCCAA CCTTTAACGT CGGATGGCCG CGAGACGGCA CCTTTAACCG 1321 AGACCTCATC ACCCAGGTTA AGATCAAGGT CTTTTCACCT GGCCCGCATG GACACCCAGA 1381 CCAGGTCCCC TACATCGTGA CCTGGGAAGC CTTGGCTTTT GACCCCCCTC CCTGGGTCAA 1441 GCCCTTTGTA CACCCTAAGC CTCCGCCTCC TCTTCCTCCA TCCGCCCCGT CTCTCCCCCT 1501 TGAACCTCCT CGTTCGACCC CGCCTCGATC CTCCCTTTAT CCAGCCCTCA CTCCTTCTCT 1561 AGGCGCCGGA ATTGAAGATC ATAGTTATTA ATAGTAATCA ATTACGGGGT CATTAGTTCA 1621 TAGCCCATAT ATGGAGTTCC GCGTTACATA ACTTACGGTA AATGGCCCGC CTGGCTGACC 1681 GCCCAACGAC CCCCGCCCAT TGACGTCAAT AATGACGTAT GTTCCCATAG TAACGCCAAT 1741 AGGGACTTTC CATTGACGTC AATGGGTGGA GTATTTACGG TAAACTGCCC ACTTGGCAGT 1801 ACATCAAGTG TATCATATGC CAAGTACGCC CCCTATTGAC GTCAATGACG GTAAATGGCC 1861 CGCCTGGCAT TATGCCCAGT ACATGACCTT ATGGGACTTT CCTACTTGGC AGTACATCTA 1921 CGTATTAGTC ATCGCTATTA CCATGGTGAT GCGGTTTTGG CAGTACATCA ATGGGCGTGG 1981 ATAGCGGTTT GACTCACGGG GATTTCCAAG TCTCCACCCC ATTGACGTCA ATGGGAGTTT 2041 GTTTTGGCAC CAAAATCAAC GGGACTTTCC AAAATGTCGT AACAACTCCG CCCCATTGAC 2101 GCAAATGGGC GGTAGGCGTG TACGGTGGGA GGTCTATATA AGCAGAGCTG GTTTAGTGAA 2161 CCGTCAGATC CGCTAGCGCT ACCGGACTCA GATCTCGAGC TCAAGCTTCG AATTCTGCAG 2221 TCGACGGTAC CGCGGGCCCG GGATCCACCG GTCATGGTGA GCAAGGGCGC CGAGCTGTTC 2281 ACCGGCATCG TGCCCATCCT GATCGAGCTG AATGGCGATG TGAATGGCCA CAAGTTCAGC 2341 GTGAGCGGCG AGGGCGAGGG CGATGCCACC TACGGCAAGC TGACCCTGAA GTTCATCTGC 2401 ACCACCGGCA AGCTGCCTGT GCCCTGGCCC ACCCTGGTGA CCACCCTGAG CTACGGCGTG 2461 CAGTGCTTCT CACGCTACCC CGATCACATG AAGCAGCACG ACTTCTTCAA GAGCGCCATG 2521 CCTGAGGGCT ACATCCAGGA GCGCACCATC TTCTTCGAGG ATGACGGCAA CTACAAGTCG 2581 CGCGCCGAGG TGAAGTTCGA GGGCGATACC CTGGTGAATC GCATCGAGCT GACCGGCACC 2641 GATTTCAAGG AGGATGGCAA CATCCTGGGC AATAAGATGG AGTACAACTA CAACGCCCAC 2701 AATGTGTACA TCATGACCGA CAAGGCCAAG AATGGCATCA AGGTGAACTT CAAGATCCGC 2761 CACAACATCG AGGATGGCAG CGTGCAGCTG GCCGACCACT ACCAGCAGAA TACCCCCATC 2821 GGCGATGGCC CTGTGCTGCT GCCCGATAAC CACTACCTGT CCACCCAGAG CGCCCTGTCC 2881 AAGGACCCCA ACGAGAAGCG CGATCACATG ATCTACTTCG GCTTCGTGAC CGCCGCCGCC 2941 ATCACCCACG GCATGGATGA GCTGTACAAG TGAGCGGCCG CGACTCTAGA TAATTCTACC 3001 GGGTAGGGGA GGCGCTTTTC CCAAGGCAGT CTGGAGCATG CGCTTTAGCA GCCCCGCTGG 3061 GCACTTGGCG CTACACAAGT GGCCTCTGGC CTCGCACACA TTCCACATCC ACCGGTAGGC 3121 GCCAACCGGC TCCGTTCTTT GGTGGCCCCT TCGCGCCACC TTCTACTCCT CCCCTAGTCA 3181 GGAAGTTCCC CCCCGCCCCG CAGCTCGCGT CGTGCAGGAC GTGACAAATG GAAGTAGCAC 3241 GTCTCACTAG TCTCGTGCAG ATGGACAGCA CCGCTGAGCA ATGGAAGCGG GTAGGCCTTT 3301 GGGGCAGCGG CCAATAGCAG CTTTGCTCCT TCGCTTTCTG GGCTCAGAGG CTGGGAAGGG 3361 GTGGGTCCGG GGGCGGGCTC AGGGGCGGGC TCAGGGGCGG GGCGGGCGCC CGAAGGTCCT 3421 CCGGAGGCCC GGCATTCTGC ACGCTTCAAA AGCGCACGTC TGCCGCGCTG TTCTCCTCTT 3481 CCTCATCTCC GGGCCTTTCG ACCTGCAGCC CAAGCTTACC ATGACCGAGT ACAAGCCCAC 3541 GGTGCGCCTC GCCACCCGCG ACGACGTCCC CAGGGCCGTA CGCACCCTCG CCGCCGCGTT 3601 CGCCGACTAC CCCGCCACGC GCCACACCGT CGATCCGGAC CGCCACATCG AGCGGGTCAC 3661 CGAGCTGCAA GAACTCTTCC TCACGCGCGT CGGGCTCGAC ATCGGCAAGG TGTGGGTCGC 3721 GGACGACGGC GCCGCGGTGG CGGTCTGGAC CACGCCGGAG AGCGTCGAAG CGGGGGCGGT 3781 GTTCGCCGAG ATCGGCCCGC GCATGGCCGA GTTGAGCGGT TCCCGGCTGG CCGCGCAGCA 3841 ACAGATGGAA GGCCTCCTGG CGCCGCACCG GCCCAAGGAG CCCGCGTGGT TCCTGGCCAC 3901 CGTCGGCGTC TCGCCCGACC ACCAGGGCAA GGGTCTGGGC AGCGCCGTCG TGCTCCCCGG 3961 AGTGGAGGCG GCCGAGCGCG CCGGGGTGCC CGCCTTCCTG GAGACCTCCG CGCCCCGCAA 4021 CCTCCCCTTC TACGAGCGGC TCGGCTTCAC CGTCACCGCC GACGTCGAGG TGCCCGAAGG 4081 ACCGCGCACC TGGTGCATGA CCCGCAAGCC CGGTGCCTGA CGCCCGCCCC ACGACCCGCA

上海泽叶生物科技有限公司
实名认证
钻石会员
入驻年限:9年